Protein-coding gene in the species Homo sapiens
| ZEB2 |
|---|
 |
| Available structures |
|---|
| PDB | Ortholog search: PDBe RCSB |
|---|
|
|
| Identifiers |
|---|
| Aliases | ZEB2, HSPC082, SIP-1, SIP1, SMADIP1, ZFHX1B, zinc finger E-box binding homeobox 2 |
|---|
| External IDs | OMIM: 605802 MGI: 1344407 HomoloGene: 8868 GeneCards: ZEB2 |
|---|
| Gene location (Human) |
|---|
 | | Chr. | Chromosome 2 (human)[1] |
|---|
| | Band | 2q22.3 | Start | 144,364,364 bp[1] |
|---|
| End | 144,521,057 bp[1] |
|---|
|
| Gene location (Mouse) |
|---|
 | | Chr. | Chromosome 2 (mouse)[2] |
|---|
| | Band | 2|2 B | Start | 44,873,644 bp[2] |
|---|
| End | 45,007,407 bp[2] |
|---|
|
| RNA expression pattern |
|---|
| Bgee | | Human | Mouse (ortholog) |
|---|
| Top expressed in | - sural nerve
- monocyte
- Achilles tendon
- corpus callosum
- internal globus pallidus
- inferior ganglion of vagus nerve
- amygdala
- external globus pallidus
- right coronary artery
- putamen
|
| | Top expressed in | - belly cord
- dentate gyrus
- iris
- Region I of hippocampus proper
- body of femur
- ankle
- olfactory bulb
- subcutaneous adipose tissue
- ganglionic eminence
- ciliary body
|
| | More reference expression data |
|
|---|
| BioGPS |  | | More reference expression data |
|
|---|
|
| Gene ontology |
|---|
| Molecular function | - DNA binding
- phosphatase regulator activity
- metal ion binding
- protein binding
- nucleic acid binding
- DNA-binding transcription repressor activity, RNA polymerase II-specific
- DNA-binding transcription activator activity, RNA polymerase II-specific
- R-SMAD binding
- sequence-specific DNA binding
- DNA-binding transcription factor activity, RNA polymerase II-specific
| | Cellular component | - nucleus
- nucleolus
- cytosol
| | Biological process | - regulation of transcription, DNA-templated
- nervous system development
- transcription, DNA-templated
- positive regulation of melanin biosynthetic process
- negative regulation of transcription by RNA polymerase II
- neural crest cell migration
- somitogenesis
- neural tube closure
- transcription by RNA polymerase II
- central nervous system development
- corpus callosum morphogenesis
- hippocampus development
- cell proliferation in forebrain
- corticospinal tract morphogenesis
- positive regulation of Wnt signaling pathway
- positive regulation of JUN kinase activity
- positive regulation of melanocyte differentiation
- positive regulation of transcription by RNA polymerase II
- developmental pigmentation
- embryonic morphogenesis
- collateral sprouting
- positive regulation of axonogenesis
- mammillary axonal complex development
- melanocyte migration
- positive regulation of lens fiber cell differentiation
- regulation of melanosome organization
| | Sources:Amigo / QuickGO |
|
| Orthologs |
|---|
| Species | Human | Mouse |
|---|
| Entrez | | |
|---|
| Ensembl | | |
|---|
| UniProt | | |
|---|
| RefSeq (mRNA) | | NM_015753
NM_001289521
NM_001355288
NM_001355289
NM_001355290
|
|---|
NM_001355291 |
|
|---|
| RefSeq (protein) | | NP_001276450
NP_056568
NP_001342217
NP_001342218
NP_001342219
|
|---|
NP_001342220 |
|
|---|
| Location (UCSC) | Chr 2: 144.36 – 144.52 Mb | Chr 2: 44.87 – 45.01 Mb |
|---|
| PubMed search | [3] | [4] |
|---|
|
| Wikidata |
| View/Edit Human | View/Edit Mouse |
|
Zinc finger E-box-binding homeobox 2 is a protein that in humans is encoded by the ZEB2 gene.[5] The ZEB2 protein is a transcription factor that plays a role in the transforming growth factor β (TGFβ) signaling pathways that are essential during early fetal development.[6]
Function
ZEB2 (previously also known as SMADIP1, SIP1) and its mammalian paralog ZEB1 belongs to the Zeb family within the ZF (zinc finger) class of homeodomain transcription factors. ZEB2 protein has 8 zinc fingers and 1 homeodomain.[7] The structure of the homeodomain shown on the right.
ZEB2 interacts with receptor-mediated, activated full-length SMADs.[5] The activation of TGFβ receptors brings about the phosphorylation of intracellular effector molecules, R-SMADs. ZEB2 is an R-SMAD-binding protein and acts as a transcriptional corepressor. It is involved in the timing of the conversion of neuroepithelial cells into radial glial cells in early development, a mechanism thought to allow for the large differences in brain size between humans and other mammals.[8]
ZEB2 transcripts are found in tissues differentiated from the neural crest such as the cranial nerve ganglia, dorsal root ganglia, sympathetic ganglionic chains, the enteric nervous system and melanocytes. ZEB2 is also found in tissues that are not derived from the neural crest, including the wall of the digestive tract, kidneys, and skeletal muscles.
Clinical significance
Mutations in the ZEB2 gene are associated with the Mowat–Wilson syndrome. This disease exhibits mutations and even complete deletions of the ZEB2 gene. Mutations of the gene can cause the gene to produce nonfunctional ZEB2 proteins or inactivate the function of the gene as a whole. These deficits of ZEB2 protein interfere with the development of many organs. Many of the symptoms can be explained by the irregular development of the structures from the neural crest.[9]
Hirschsprung's disease also has many symptoms that can be explained by lack of ZEB2 during development of the digestive tract nerves. This disease causes severe constipation and enlargement of the colon.[10]
The risk of hepatocellular carcinoma and cirrhosis in chronic hepatitis B has been reported to be associated with a single-nucleotide polymorphism in the promoter region of ZEB2, designated rs3806475, under a recessive model of inheritance.[11]
References
- ^ a b c GRCh38: Ensembl release 89: ENSG00000169554 – Ensembl, May 2017
- ^ a b c GRCm38: Ensembl release 89: ENSMUSG00000026872 – Ensembl, May 2017
- ^ "Human PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- ^ "Mouse PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- ^ a b "Entrez Gene: ZEB2 zinc finger E-box binding homeobox 2".
- ^ Bassez G, Camand OJ, Cacheux V, Kobetz A, Dastot-Le Moal F, Marchant D, Catala M, Abitbol M, Goossens M (March 2004). "Pleiotropic and diverse expression of ZFHX1B gene transcripts during mouse and human development supports the various clinical manifestations of the "Mowat-Wilson" syndrome". Neurobiology of Disease. 15 (2): 240–50. doi:10.1016/j.nbd.2003.10.004. PMID 15006694. S2CID 25770329.
- ^ Bürglin TR, Affolter M (June 2016). "Homeodomain proteins: an update". Chromosoma. 125 (3): 497–521. doi:10.1007/s00412-015-0543-8. PMC 4901127. PMID 26464018.
- ^ Benito-Kwiecinski, Silvia; Giandomenico, Stefano L.; Sutcliffe, Magdalena; Riis, Erlend S.; Freire-Pritchett, Paula; Kelava, Iva; Wunderlich, Stephanie; Martin, Ulrich; Wray, Gregory A.; McDole, Kate; Lancaster, Madeline A. (2021). "An early cell shape transition drives evolutionary expansion of the human forebrain". Cell. 184 (8): 2084–2102.e19. doi:10.1016/j.cell.2021.02.050. PMC 8054913. PMID 33765444.
- ^ Dastot-Le Moal F, Wilson M, Mowat D, Collot N, Niel F, Goossens M (April 2007). "ZFHX1B mutations in patients with Mowat-Wilson syndrome". Human Mutation. 28 (4): 313–21. doi:10.1002/humu.20452. PMID 17203459. S2CID 37981110.
- ^ Saunders CJ, Zhao W, Ardinger HH (November 2009). "Comprehensive ZEB2 gene analysis for Mowat-Wilson syndrome in a North American cohort: a suggested approach to molecular diagnostics". American Journal of Medical Genetics Part A. 149A (11): 2527–31. doi:10.1002/ajmg.a.33067. PMID 19842203. S2CID 22472646.
- ^ Liu WX, Yang L, Yan HM, Yan LN, Zhang XL, Ma N, Tang LM, Gao X, Liu DW (2021). Argentiero A, Mehta R, Wang S (eds.). "Germline Variants and Genetic Interactions of Several EMT Regulatory Genes Increase the Risk of HBV-Related Hepatocellular Carcinoma". Frontiers in Oncology. 11. doi:10.3389/fonc.2021.564477. PMC 8226114. PMID 34178612.
 This article incorporates text from this source, which is available under the CC BY 4.0 license.
This article incorporates text from this source, which is available under the CC BY 4.0 license.
Further reading
- Mowat DR, Wilson MJ, Goossens M (May 2003). "Mowat-Wilson syndrome". Journal of Medical Genetics. 40 (5): 305–10. doi:10.1136/jmg.40.5.305. PMC 1735450. PMID 12746390.
- Nagase T, Ishikawa K, Miyajima N, Tanaka A, Kotani H, Nomura N, Ohara O (February 1998). "Prediction of the coding sequences of unidentified human genes. IX. The complete sequences of 100 new cDNA clones from brain which can code for large proteins in vitro". DNA Research. 5 (1): 31–9. doi:10.1093/dnares/5.1.31. PMID 9628581.
- Ueki N, Oda T, Kondo M, Yano K, Noguchi T, Muramatsu M (December 1998). "Selection system for genes encoding nuclear-targeted proteins". Nature Biotechnology. 16 (13): 1338–42. doi:10.1038/4315. PMID 9853615. S2CID 20001769.
- Verschueren K, Remacle JE, Collart C, Kraft H, Baker BS, Tylzanowski P, Nelles L, Wuytens G, Su MT, Bodmer R, Smith JC, Huylebroeck D (July 1999). "SIP1, a novel zinc finger/homeodomain repressor, interacts with Smad proteins and binds to 5'-CACCT sequences in candidate target genes". The Journal of Biological Chemistry. 274 (29): 20489–98. doi:10.1074/jbc.274.29.20489. PMID 10400677.
- Wakamatsu N, Yamada Y, Yamada K, Ono T, Nomura N, Taniguchi H, Kitoh H, Mutoh N, Yamanaka T, Mushiake K, Kato K, Sonta S, Nagaya M (April 2001). "Mutations in SIP1, encoding Smad interacting protein-1, cause a form of Hirschsprung disease". Nature Genetics. 27 (4): 369–70. doi:10.1038/86860. PMID 11279515. S2CID 39070888.
- Comijn J, Berx G, Vermassen P, Verschueren K, van Grunsven L, Bruyneel E, Mareel M, Huylebroeck D, van Roy F (June 2001). "The two-handed E box binding zinc finger protein SIP1 downregulates E-cadherin and induces invasion". Molecular Cell. 7 (6): 1267–78. doi:10.1016/S1097-2765(01)00260-X. PMID 11430829.
- Cacheux V, Dastot-Le Moal F, Kääriäinen H, Bondurand N, Rintala R, Boissier B, Wilson M, Mowat D, Goossens M (July 2001). "Loss-of-function mutations in SIP1 Smad interacting protein 1 result in a syndromic Hirschsprung disease". Human Molecular Genetics. 10 (14): 1503–10. doi:10.1093/hmg/10.14.1503. PMID 11448942.
- Tylzanowski P, Verschueren K, Huylebroeck D, Luyten FP (October 2001). "Smad-interacting protein 1 is a repressor of liver/bone/kidney alkaline phosphatase transcription in bone morphogenetic protein-induced osteogenic differentiation of C2C12 cells". The Journal of Biological Chemistry. 276 (43): 40001–7. doi:10.1074/jbc.M104112200. PMID 11477103.
- Yamada K, Yamada Y, Nomura N, Miura K, Wakako R, Hayakawa C, Matsumoto A, Kumagai T, Yoshimura I, Miyazaki S, Kato K, Sonta S, Ono H, Yamanaka T, Nagaya M, Wakamatsu N (December 2001). "Nonsense and frameshift mutations in ZFHX1B, encoding Smad-interacting protein 1, cause a complex developmental disorder with a great variety of clinical features". American Journal of Human Genetics. 69 (6): 1178–85. doi:10.1086/324343. PMC 1235530. PMID 11592033.
- Amiel J, Espinosa-Parrilla Y, Steffann J, Gosset P, Pelet A, Prieur M, Boute O, Choiset A, Lacombe D, Philip N, Le Merrer M, Tanaka H, Till M, Touraine R, Toutain A, Vekemans M, Munnich A, Lyonnet S (December 2001). "Large-scale deletions and SMADIP1 truncating mutations in syndromic Hirschsprung disease with involvement of midline structures". American Journal of Human Genetics. 69 (6): 1370–7. doi:10.1086/324342. PMC 1235547. PMID 11595972.
- Zweier C, Albrecht B, Mitulla B, Behrens R, Beese M, Gillessen-Kaesbach G, Rott HD, Rauch A (March 2002). ""Mowat-Wilson" syndrome with and without Hirschsprung disease is a distinct, recognizable multiple congenital anomalies-mental retardation syndrome caused by mutations in the zinc finger homeo box 1B gene". American Journal of Medical Genetics. 108 (3): 177–81. doi:10.1002/ajmg.10226. PMID 11891681.
- Nagaya M, Kato J, Niimi N, Tanaka S, Wakamatsu N (August 2002). "Clinical features of a form of Hirschsprung's disease caused by a novel genetic abnormality". Journal of Pediatric Surgery. 37 (8): 1117–22. doi:10.1053/jpsu.2002.34455. PMID 12149685.
- Guaita S, Puig I, Franci C, Garrido M, Dominguez D, Batlle E, Sancho E, Dedhar S, De Herreros AG, Baulida J (October 2002). "Snail induction of epithelial to mesenchymal transition in tumor cells is accompanied by MUC1 repression and ZEB1 expression". The Journal of Biological Chemistry. 277 (42): 39209–16. doi:10.1074/jbc.M206400200. PMID 12161443.
- Espinosa-Parrilla Y, Amiel J, Augé J, Encha-Razavi F, Munnich A, Lyonnet S, Vekemans M, Attié-Bitach T (June 2002). "Expression of the SMADIP1 gene during early human development". Mechanisms of Development. 114 (1–2): 187–91. doi:10.1016/S0925-4773(02)00062-X. PMID 12175509. S2CID 18645909.
- Yoneda M, Fujita T, Yamada Y, Yamada K, Fujii A, Inagaki T, Nakagawa H, Shimada A, Kishikawa M, Nagaya M, Azuma T, Kuriyama M, Wakamatsu N (November 2002). "Late infantile Hirschsprung disease-mental retardation syndrome with a 3-bp deletion in ZFHX1B". Neurology. 59 (10): 1637–40. doi:10.1212/01.wnl.0000034842.78350.4e. PMID 12451214. S2CID 34389990.
- Postigo AA (May 2003). "Opposing functions of ZEB proteins in the regulation of the TGFbeta/BMP signaling pathway". The EMBO Journal. 22 (10): 2443–52. doi:10.1093/emboj/cdg225. PMC 155983. PMID 12743038.
- Postigo AA, Depp JL, Taylor JJ, Kroll KL (May 2003). "Regulation of Smad signaling through a differential recruitment of coactivators and corepressors by ZEB proteins". The EMBO Journal. 22 (10): 2453–62. doi:10.1093/emboj/cdg226. PMC 155984. PMID 12743039.
- Zweier C, Temple IK, Beemer F, Zackai E, Lerman-Sagie T, Weschke B, Anderson CE, Rauch A (August 2003). "Characterisation of deletions of the ZFHX1B region and genotype-phenotype analysis in Mowat-Wilson syndrome". Journal of Medical Genetics. 40 (8): 601–5. doi:10.1136/jmg.40.8.601. PMC 1735564. PMID 12920073.
External links
- GeneReviews/NIH/NCBI/UW entry on Mowat-Wilson syndrome
- ZEB2+protein,+human at the U.S. National Library of Medicine Medical Subject Headings (MeSH)
|
|---|
(1) Basic domains |
|---|
| (1.1) Basic leucine zipper (bZIP) | |
|---|
| (1.2) Basic helix-loop-helix (bHLH) | | Group A | |
|---|
| Group B | |
|---|
Group C
bHLH-PAS | |
|---|
| Group D | |
|---|
| Group E | |
|---|
Group F
bHLH-COE | |
|---|
|
|---|
| (1.3) bHLH-ZIP | |
|---|
| (1.4) NF-1 | |
|---|
| (1.5) RF-X | |
|---|
| (1.6) Basic helix-span-helix (bHSH) | |
|---|
|
|
(2) Zinc finger DNA-binding domains |
|---|
| (2.1) Nuclear receptor (Cys4) | | subfamily 1 | |
|---|
| subfamily 2 | |
|---|
| subfamily 3 | |
|---|
| subfamily 4 | |
|---|
| subfamily 5 | |
|---|
| subfamily 6 | |
|---|
| subfamily 0 | |
|---|
|
|---|
| (2.2) Other Cys4 | |
|---|
| (2.3) Cys2His2 | |
|---|
| (2.4) Cys6 | |
|---|
| (2.5) Alternating composition | |
|---|
| (2.6) WRKY | |
|---|
|
|
|
(4) β-Scaffold factors with minor groove contacts |
|---|
|
|
(0) Other transcription factors |
|---|
|
|
see also transcription factor/coregulator deficiencies |

 This article incorporates text from this source, which is available under the CC BY 4.0 license.
This article incorporates text from this source, which is available under the CC BY 4.0 license. 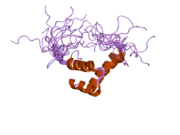 2da7: Solution structure of the homeobox domain of Zinc finger homeobox protein 1b (Smad interacting protein 1)
2da7: Solution structure of the homeobox domain of Zinc finger homeobox protein 1b (Smad interacting protein 1)




















