Protein-coding gene in humans
| BTC |
|---|
|
| Available structures |
|---|
| PDB | Ortholog search: PDBe RCSB |
|---|
| List of PDB id codes |
|---|
1IOX, 1IP0 |
|
|
| Identifiers |
|---|
| Aliases | BTC, entrez:685, betacellulin |
|---|
| External IDs | OMIM: 600345; MGI: 99439; HomoloGene: 1309; GeneCards: BTC; OMA:BTC - orthologs |
|---|
| Gene location (Human) |
|---|
 | | Chr. | Chromosome 4 (human)[1] |
|---|
| | Band | 4q13.3 | Start | 74,744,759 bp[1] |
|---|
| End | 74,794,523 bp[1] |
|---|
|
| Gene location (Mouse) |
|---|
 | | Chr. | Chromosome 5 (mouse)[2] |
|---|
| | Band | 5 E2|5 44.78 cM | Start | 91,505,120 bp[2] |
|---|
| End | 91,550,853 bp[2] |
|---|
|
| RNA expression pattern |
|---|
| Bgee | | Human | Mouse (ortholog) |
|---|
| Top expressed in | - gastric mucosa
- bronchial epithelial cell
- ascending aorta
- retinal pigment epithelium
- right coronary artery
- skin of abdomen
- rectum
- popliteal artery
- transverse colon
- gallbladder
|
| | Top expressed in | - mucous cell of stomach
- pyloric antrum
- epithelium of stomach
- medullary collecting duct
- skin of back
- retinal pigment epithelium
- cumulus cell
- skin of abdomen
- duodenum
- belly cord
|
| | More reference expression data |
|
|---|
| BioGPS | |
|---|
|
| Gene ontology |
|---|
| Molecular function | - protein binding
- phosphatidylinositol-4,5-bisphosphate 3-kinase activity
- protein tyrosine kinase activity
- epidermal growth factor receptor binding
- growth factor activity
| | Cellular component | - integral component of membrane
- membrane
- plasma membrane
- extracellular region
- intracellular anatomical structure
- extracellular space
- clathrin-coated vesicle membrane
| | Biological process | - positive regulation of urine volume
- positive regulation of fibroblast proliferation
- negative regulation of apoptotic process
- MAPK cascade
- positive regulation of cell differentiation
- positive regulation of cell population proliferation
- positive regulation of mitotic nuclear division
- positive regulation of cell division
- regulation of cell motility
- phosphatidylinositol phosphate biosynthetic process
- epidermal growth factor receptor signaling pathway
- ERBB2 signaling pathway
- peptidyl-tyrosine phosphorylation
- regulation of signaling receptor activity
- positive regulation of protein kinase B signaling
- signal transduction
- negative regulation of epidermal growth factor receptor signaling pathway
- membrane organization
| | Sources:Amigo / QuickGO |
|
|
| Wikidata |
| View/Edit Human | View/Edit Mouse |
|
Betacellulin is a protein that in humans is encoded by the BTC gene located on chromosome 4 at locus 4q13-q21.[5] Betacellulin was initially identified as a mitogen.[6] Betacellulin, is a part of an Epidermal Growth Factor (EGF) family and functions as a ligand for the epidermal growth factor receptor (EGFR). The role of betacellulin as an EGF is manifested differently in various tissues, and it has a great effect on nitrogen signaling in retinal pigment epithelial cells and vascular smooth muscle cells. While many studies attest a role for betacellulin in the differentiation of pancreatic β-cells, the last decade witnessed the association of betacellulin with many additional biological processes, ranging from reproduction to the control of neural stem cells.[6] Betacellulin is a member of the EGF family of growth factors. It is synthesized primarily as a transmembrane precursor, which is then processed to mature molecule by proteolytic events.
Structure
As shown on figure 1, the secondary structure of the human betacellulin-2 has 6% helical (1 helices; 3 residues) 36% beta sheet (5 strands; 18 residues). The mRNA of betacellulin contains six exons in which is 2816 base-pair long.[6] The mRNA was translated into 178 amino acids, and different regions of the amino acid are responsible for different function.[6] The first 31 amino acids are responsible for the signal peptide (Figure 2, exon 1), the 32nd to 118th amino acids are responsible for the extracellular region (Figure 2, exon 2 and 3), the 65-105 amino acids are responsible for the EGF-like domain (Figure 2, exon 3), the transmembrane domain is from amino acids 119-139 (Figure 2, exon 4), the cytoplasmic tail is from amino acid 140-178 (Figure 2, exon 5).[6]
Function
As a typical EGFR ligand, betacellulin is expressed by a variety of cell types and tissues, the post-translation of the betacellulin can ectodomain shedding, and the proteolytic release the soluble factors can bind and activate the homodimer or heterodimer of the ERBB receptors. The membrane-anchored form of the betacellulin can activate the epidermal growth factor receptor (EGFR).[7] Betacellulin stimulates the proliferation of retinal pigment epithelial and vascular smooth muscle cells but did not stimulate the growth of several other cell types, such as endothelial cells and fetal lung fibroblasts.[8]
Tissue distribution
The mRNA coding for betacellulin was found to be slightly higher compared in the rat sciatic nerve segment after nerve damage, suggesting that betacellulin can play a role in peripheral nerve regeneration. Immunohistochemistry has been used to look for betacellulin expression in Schwann cells. Treating cells with betacellulin recombinant protein can be used to investigate the role of betacellulin in managing Schwann cells. A co-culture assay can also used to assess the effect of Schwann cell-secreted betacellulin on neurons.[9]
Mouse BTC is expressed as a 178-amino acid precursor. The membrane-bound precursor is cleaved to yield mature secreted mouse BTC. BTC is synthesized in a wide range of adult tissues and in many cultured cells, including smooth muscle cells and epithelial cells. The amino acid sequence of mature mouse BTC is 82.5%, identical with that of human BTC, and both exhibit significant overall similarity with other members of the EGF family.
Clinical significance
The transcription factor signal transducer and activator of transcription 3 (STAT3) was identified as the therapeutic target for glioblastoma.[10]
References
- ^ a b c GRCh38: Ensembl release 89: ENSG00000174808 – Ensembl, May 2017
- ^ a b c GRCm38: Ensembl release 89: ENSMUSG00000082361 – Ensembl, May 2017
- ^ "Human PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- ^ "Mouse PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- ^ "Entrez Gene: betacellulin".
- ^ a b c d e Dahlhoff M, Wolf E, Schneider MR (April 2014). "The ABC of BTC: structural properties and biological roles of betacellulin". Seminars in Cell & Developmental Biology. 28: 42–48. doi:10.1016/j.semcdb.2014.01.002. PMID 24440602.
- ^ Dahlhoff M, Wolf E, Schneider MR (April 2014). "The ABC of BTC: structural properties and biological roles of betacellulin". Seminars in Cell & Developmental Biology. 28: 42–48. doi:10.1016/j.semcdb.2014.01.002. PMID 24440602.
- ^ Sethi G, Ahn KS, Chaturvedi MM, Aggarwal BB (15 November 2007). "Epidermal growth factor (EGF) activates nuclear factor-κB through IκBα kinase-independent but EGF receptor-kinase dependent tyrosine 42 phosphorylation of IκBα". Oncogene. 26 (52): 7324–7332. doi:10.1038/sj.onc.1210544. PMID 17533369. S2CID 25253064.
- ^ Wang Y, Zhang F, Zhang Y, Shan Q, Liu W, Zhang F, et al. (March 2021). "Betacellulin regulates peripheral nerve regeneration by affecting Schwann cell migration and axon elongation". Molecular Medicine. 27 (1): 27. doi:10.1186/s10020-021-00292-5. PMC 8015203. PMID 33794764.
- ^ Fan Q, An Z, Wong RA, Luo X, Lu ED, Baldwin A, et al. (April 2020). "Betacellulin drives therapy resistance in glioblastoma". Neuro-Oncology. 22 (4): 457–469. doi:10.1093/neuonc/noz206. PMC 7158663. PMID 31678994.
Further reading
- Kim HS, Shin HS, Kwak HJ, Cho CH, Lee CO, Koh GY (February 2003). "Betacellulin induces angiogenesis through activation of mitogen-activated protein kinase and phosphatidylinositol 3'-kinase in endothelial cell". FASEB Journal. 17 (2): 318–320. doi:10.1096/fj.02-0570fje. PMID 12475887. S2CID 25722265.
- Yamamoto T, Akisue T, Marui T, Nakatani T, Kawamoto T, Hitora T, et al. (2004). "Expression of betacellulin, heparin-binding epidermal growth factor and epiregulin in human malignant fibrous histiocytoma". Anticancer Research. 24 (3b): 2007–2010. PMID 15274392.
- Nakagawa T, Furuta H, Sanke T, Sakagashira S, Shimomura H, Shimajiri Y, et al. (June 2005). "Molecular scanning of the betacellulin gene for mutations in type 2 diabetic patients". Diabetes Research and Clinical Practice. 68 (3): 188–192. doi:10.1016/j.diabres.2004.09.019. PMID 15936459.
- Silver K, Tolea M, Wang J, Pollin TI, Yao F, Mitchell BD (April 2005). "The exon 1 Cys7Gly polymorphism within the betacellulin gene is associated with type 2 diabetes in African Americans". Diabetes. 54 (4): 1179–1184. doi:10.2337/diabetes.54.4.1179. PMID 15793259.
- Tanimura K, Nakago S, Murakoshi H, Takekida S, Moriyama T, Matsuo H, et al. (July 2004). "Changes in the expression and cytological localization of betacellulin and its receptors (ErbB-1 and ErbB-4) in the trophoblasts in human placenta over the course of pregnancy". European Journal of Endocrinology. 151 (1): 93–101. doi:10.1530/eje.0.1510093. hdl:20.500.14094/D1003166. PMID 15248827.
- Saito T, Okada S, Ohshima K, Yamada E, Sato M, Uehara Y, et al. (September 2004). "Differential activation of epidermal growth factor (EGF) receptor downstream signaling pathways by betacellulin and EGF". Endocrinology. 145 (9): 4232–4243. doi:10.1210/en.2004-0401. PMID 15192046.
- Elbein SC, Wang X, Karim MA, Chu WS, Silver KD (July 2006). "Analysis of coding variants in the betacellulin gene in type 2 diabetes and insulin secretion in African American subjects". BMC Medical Genetics. 7: 62. doi:10.1186/1471-2350-7-62. PMC 1544326. PMID 16869959.
- Genetos DC, Rao RR, Vidal MA (April 2010). "Betacellulin inhibits osteogenic differentiation and stimulates proliferation through HIF-1alpha". Cell and Tissue Research. 340 (1): 81–89. doi:10.1007/s00441-010-0929-0. PMC 2847694. PMID 20165885.
- Moss ML, Bomar M, Liu Q, Sage H, Dempsey P, Lenhart PM, et al. (December 2007). "The ADAM10 prodomain is a specific inhibitor of ADAM10 proteolytic activity and inhibits cellular shedding events". The Journal of Biological Chemistry. 282 (49): 35712–35721. doi:10.1074/jbc.M703231200. PMID 17895248.
- Rittié L, Kansra S, Stoll SW, Li Y, Gudjonsson JE, Shao Y, et al. (June 2007). "Differential ErbB1 signaling in squamous cell versus basal cell carcinoma of the skin". The American Journal of Pathology. 170 (6): 2089–2099. doi:10.2353/ajpath.2007.060537. PMC 1899432. PMID 17525275.
- Révillion F, Lhotellier V, Hornez L, Bonneterre J, Peyrat JP (January 2008). "ErbB/HER ligands in human breast cancer, and relationships with their receptors, the bio-pathological features and prognosis". Annals of Oncology. 19 (1): 73–80. doi:10.1093/annonc/mdm431. PMID 17962208.
- Silver KD, Shi X, Mitchell BD (April 2007). "Betacellulin variants and type 2 diabetes in the Old Order Amish". Experimental and Clinical Endocrinology & Diabetes. 115 (4): 229–231. doi:10.1055/s-2007-970575. PMID 17479438.
- Mehrle A, Rosenfelder H, Schupp I, del Val C, Arlt D, Hahne F, et al. (January 2006). "The LIFEdb database in 2006". Nucleic Acids Research. 34 (Database issue): D415–D418. doi:10.1093/nar/gkj139. PMC 1347501. PMID 16381901.
- Stoeck A, Shang L, Dempsey PJ (July 2010). "Sequential and gamma-secretase-dependent processing of the betacellulin precursor generates a palmitoylated intracellular-domain fragment that inhibits cell growth". Journal of Cell Science. 123 (Pt 13): 2319–2331. doi:10.1242/jcs.060830. PMC 2886747. PMID 20530572.
- Nagaoka T, Fukuda T, Hashizume T, Nishiyama T, Tada H, Yamada H, et al. (June 2008). "A betacellulin mutant promotes differentiation of pancreatic acinar AR42J cells into insulin-producing cells with low affinity of binding to ErbB1". Journal of Molecular Biology. 380 (1): 83–94. doi:10.1016/j.jmb.2008.03.054. PMID 18508082.
- Nakano Y, Furuta H, Doi A, Matsuno S, Nakagawa T, Shimomura H, et al. (December 2005). "A functional variant in the human betacellulin gene promoter is associated with type 2 diabetes". Diabetes. 54 (12): 3560–3566. doi:10.2337/diabetes.54.12.3560. PMID 16306376.
- Sanderson MP, Erickson SN, Gough PJ, Garton KJ, Wille PT, Raines EW, et al. (January 2005). "ADAM10 mediates ectodomain shedding of the betacellulin precursor activated by p-aminophenylmercuric acetate and extracellular calcium influx". The Journal of Biological Chemistry. 280 (3): 1826–1837. doi:10.1074/jbc.M408804200. PMID 15507448.
- Dunbar AJ, Goddard C (August 2000). "Structure-function and biological role of betacellulin". The International Journal of Biochemistry & Cell Biology. 32 (8): 805–815. doi:10.1016/S1357-2725(00)00028-5. PMID 10940639.
- Wiemann S, Arlt D, Huber W, Wellenreuther R, Schleeger S, Mehrle A, et al. (October 2004). "From ORFeome to biology: a functional genomics pipeline". Genome Research. 14 (10B): 2136–2144. doi:10.1101/gr.2576704. PMC 528930. PMID 15489336.
- Silver KD, Magnuson VL, Tolea M, Wang J, Hagopian WA, Mitchell BD (July 2006). "Association of a polymorphism in the betacellulin gene with type 1 diabetes mellitus in two populations". Journal of Molecular Medicine. 84 (7): 616–623. doi:10.1007/s00109-006-0052-6. PMID 16683131. S2CID 31302931.
External links
|
|---|
| Angiopoietin | - Kinase inhibitors: Altiratinib
- CE-245677
- Rebastinib
|
|---|
| CNTF | |
|---|
| EGF (ErbB) | |
|---|
| FGF | | FGFR1 | |
|---|
| FGFR2 | - Agonists: Ersofermin
- FGF (1, 2 (bFGF), 3, 4, 5, 6, 7 (KGF), 8, 9, 10 (KGF2), 17, 18, 22)
- Palifermin
- Repifermin
- Selpercatinib
- Sprifermin
- Trafermin
- Antibodies: Aprutumab
- Aprutumab ixadotin
|
|---|
| FGFR3 | |
|---|
| FGFR4 | |
|---|
| Unsorted | |
|---|
|
|---|
| HGF (c-Met) | |
|---|
| IGF | | IGF-1 | - Kinase inhibitors: BMS-754807
- Linsitinib
- NVP-ADW742
- NVP-AEW541
- OSl-906
|
|---|
| IGF-2 | - Antibodies: Dusigitumab
- Xentuzumab (against IGF-1 and IGF-2)
|
|---|
| Others | - Cleavage products/derivatives with unknown target: Glypromate (GPE, (1-3)IGF-1)
- Trofinetide
|
|---|
|
|---|
| LNGF (p75NTR) | - Aptamers: Against NGF: RBM-004
- Decoy receptors: LEVI-04 (p75NTR-Fc)
|
|---|
| PDGF | |
|---|
| RET (GFL) | |
|---|
| SCF (c-Kit) | |
|---|
| TGFβ | |
|---|
| Trk | | TrkA | - Negative allosteric modulators: VM-902A
- Aptamers: Against NGF: RBM-004
- Decoy receptors: ReN-1820 (TrkAd5)
|
|---|
| TrkB | - Agonists: 3,7-DHF
- 3,7,8,2'-THF
- 4'-DMA-7,8-DHF
- 7,3'-DHF
- 7,8-DHF
- 7,8,2'-THF
- 7,8,3'-THF
- Amitriptyline
- BDNF
- BNN-20
- Deoxygedunin
- Deprenyl
- Diosmetin
- DMAQ-B1
- HIOC
- LM22A-4
- N-Acetylserotonin
- NT-3
- NT-4
- Norwogonin (5,7,8-THF)
- R7
- R13
- TDP6
|
|---|
| TrkC | |
|---|
|
|---|
| VEGF | |
|---|
| Others | - Additional growth factors: Adrenomedullin
- Colony-stimulating factors (see here instead)
- Connective tissue growth factor (CTGF)
- Ephrins (A1, A2, A3, A4, A5, B1, B2, B3)
- Erythropoietin (see here instead)
- Glucose-6-phosphate isomerase (GPI; PGI, PHI, AMF)
- Glia maturation factor (GMF)
- Hepatoma-derived growth factor (HDGF)
- Interleukins/T-cell growth factors (see here instead)
- Leukemia inhibitory factor (LIF)
- Macrophage-stimulating protein (MSP; HLP, HGFLP)
- Midkine (NEGF2)
- Migration-stimulating factor (MSF; PRG4)
- Oncomodulin
- Pituitary adenylate cyclase-activating peptide (PACAP)
- Pleiotrophin
- Renalase
- Thrombopoietin (see here instead)
- Wnt signaling proteins
- Additional growth factor receptor modulators: Cerebrolysin (neurotrophin mixture)
|
|---|
This article incorporates text from the United States National Library of Medicine, which is in the public domain.
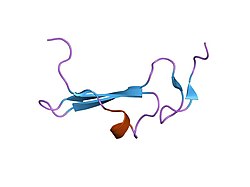 Figure 1. NMR Structure of Human Betacellulin-2
Figure 1. NMR Structure of Human Betacellulin-2 Figure 2. The transcription and translation product of betacellulin gene
Figure 2. The transcription and translation product of betacellulin gene 1iox: NMR Structure of human Betacellulin-2
1iox: NMR Structure of human Betacellulin-2 1ip0: NMR STRUCTURE OF HUMAN BETACELLULIN-2
1ip0: NMR STRUCTURE OF HUMAN BETACELLULIN-2


















